Variational Inference is hip. Therefore it might be a good idea to demonstrate that it should not be confused with a free lunch. I'll take an example from a previous post in order to demonstrate a point of weakness.
The Model
I'll continue with the chickweight model that I've defined in a previous blogpost. Here is the code for the model.
df = pd.read_csv("http://koaning.io/theme/data/chickweight.csv",
skiprows=1,
names=["r", "weight", "time", "chick", "diet"])
time_input = 10
with pm.Model() as mod:
intercept = pm.Normal("intercept", 0, 2)
time_effect = pm.Normal("time_weight_effect", 0, 2, shape=(4,))
diet = pm.Categorical("diet", p=[0.25, 0.25, 0.25, 0.25], shape=(4,), observed=dummy_rows)
sigma = pm.HalfNormal("sigma", 2)
sigma_time_effect = pm.HalfNormal("time_sigma_effect", 2, shape=(4,))
weight = pm.Normal("weight",
mu=intercept + time_effect.dot(diet.T)*df.time,
sd=sigma + sigma_time_effect.dot(diet.T)*df.time,
observed=df.weight)
trace = pm.sample(5000, chains=1)
Next I'll show how the traceplots are different if we compare different inference methods.
NUTS sampling results
I took 5500 samples with NUTS. It took about 7 seconds and this is the output:
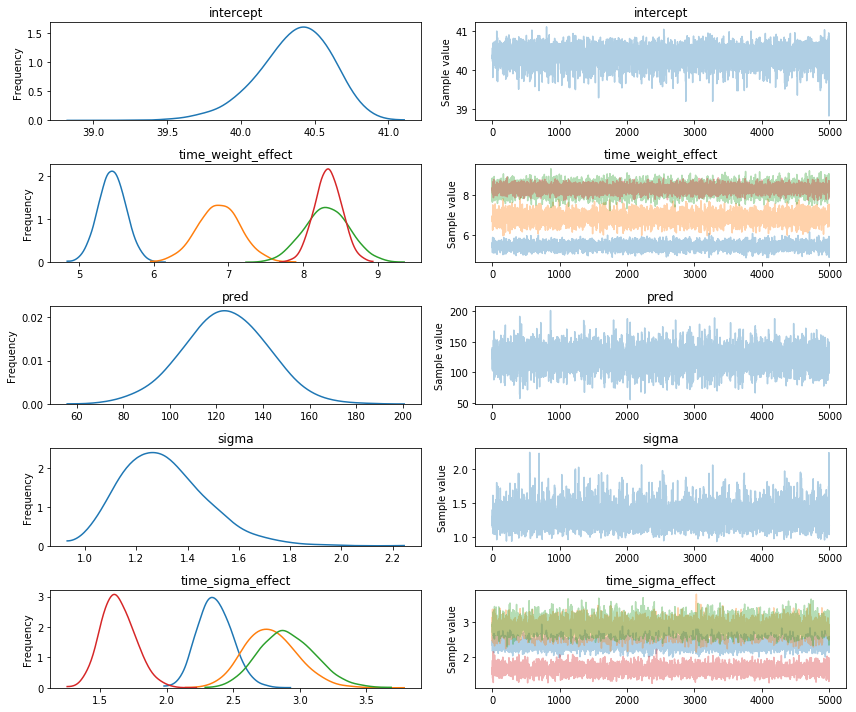
If you read the previous blogpost then you understand why this model is arguably well defined.
Metropolis sampling results
I took 20000 samples with Metropolis. It took about 14 seconds and this is the output:
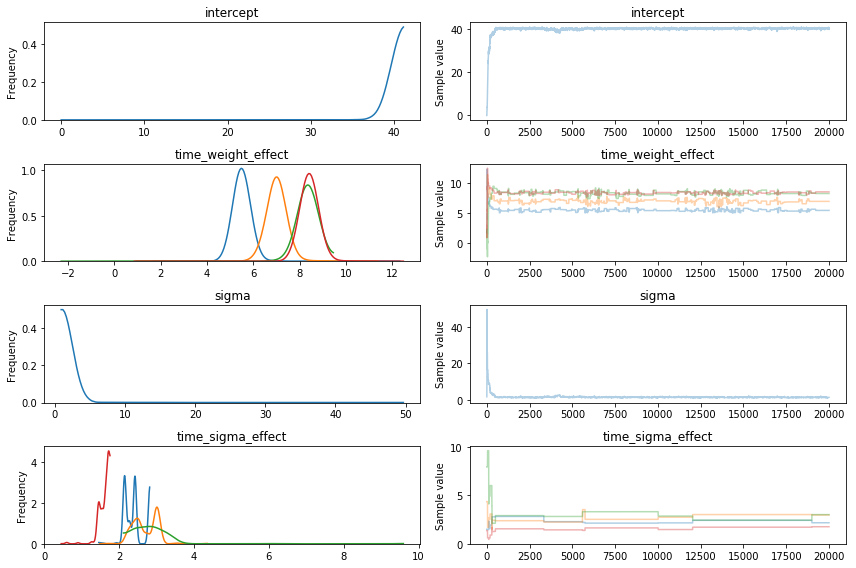
Note that the burn in isn't the only issue: the parameters that I end up with are way off.
VI results
I used the fullrank_advi setting. Here's a traceplot from the samples I took from the approximated posteriour.
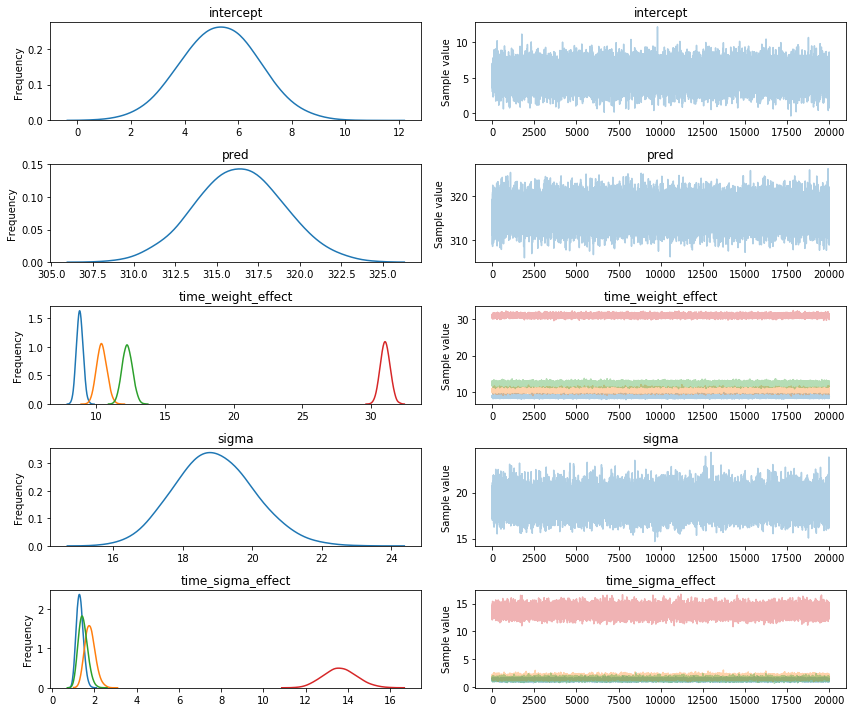
Again, the fitted estimates are nowhere near the NUTS samples.
Fix?
The interesting thing is that if I change the model slightly, VI suddenly has no issues.
n_diets = df.diet.nunique()
with pm.Model() as model:
mu_intercept = pm.Normal('mu_intercept', mu=40, sd=5)
mu_slope = pm.HalfNormal('mu_slope', 10, shape=(n_diets,))
mu = mu_intercept + mu_slope[df.diet-1] * df.time
sigma_intercept = pm.HalfNormal('sigma_intercept', sd=2)
sigma_slope = pm.HalfNormal('sigma_slope', sd=2, shape=n_diets)
sigma = sigma_intercept + sigma_slope[df.diet-1] * df.time
weight = pm.Normal('weight', mu=mu, sd=sigma, observed=df.weight)
approx = pm.fit(20000, random_seed=42, method="fullrank_advi")
The main difference is that I am no longer using pm.Categorical.
With that out of the way suddenly the estimates look a whole lot better.
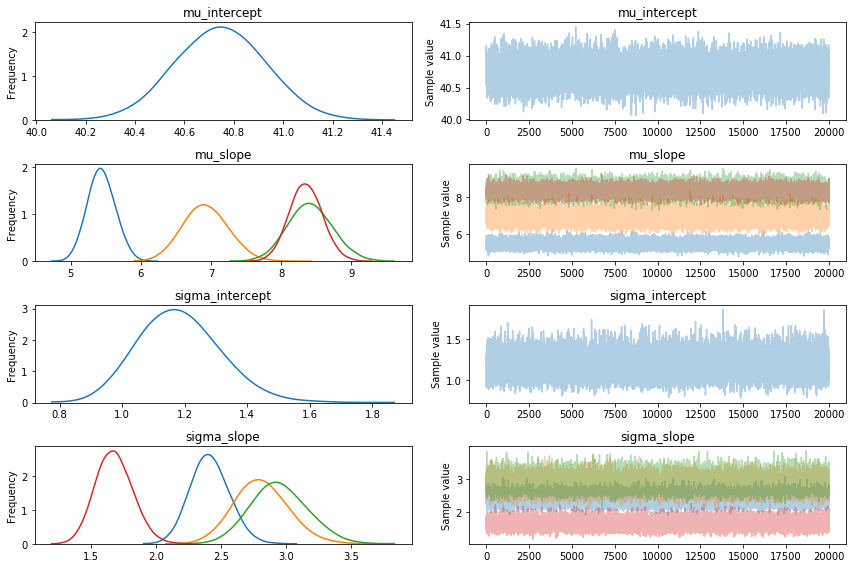
I can't accurately pinpoint what exactly is causing this massive shift but I might imagine that anything using a gradient would have trouble with something discrete in a system.
Conclusion
Be careful when using variational inference. It might be faster but it is only faster because it approximates. I'm not the only person why is a bit skeptical of variational inference.
The alternative, NUTS sampling still amazes me, even though it isn't perfect.